Project: Dimensionality reduction of white matter tracts
Description
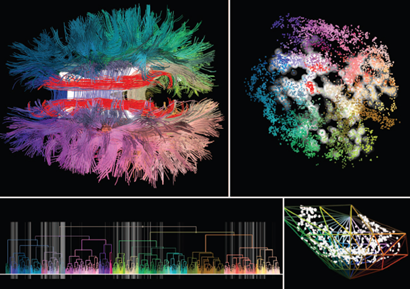
Various diffusion MRI (dMRI) measures have been proposed for characterising tissue microstructure over the last 15 years. Despite the growing number of experiments using different dMRI measures in assessments of white matter, there has been limited work on: 1) examining their covariance along specific pathways; and on 2) combining these different measures to study tissue microstructure. Indeed, it quickly becomes intractable for existing analysis pipelines to process multiple measurements at each voxel and at each vertex forming a streamline, highlighting the need for new ways to visualise or analyse such high-dimensional data.
This project aims to provide a novel visual exploration paradigm that facilitates navigation through complex fiber tracts by means of dimensionality reduction. As opposed to existing approaches solely based on tract morphology, this project will allow the navigation of microstructural features using 2D maps derived from (e.g., t-SNE, UMAP, PCA, etc.). The student will work with large, multidimensional datasets. This will ultimately allow the navigation and interaction with such dense data in a unprecedented way - allowing brain researchers to find patterns in the data - or even as a QA tool to identify outliers.
Chen, Wei, et al. "A novel interface for interactive exploration of DTI fibers." IEEE Transactions on Visualization and Computer Graphics 15.6 (2009): 1433-1440.
Jianu, Radu, Cagatay Demiralp, and David Laidlaw. "Exploring 3D DTI fiber tracts with linked 2D representations." IEEE transactions on Visualization and Computer Graphics 15.6 (2009): 1449-1456.
Franke, Loraine, et al. "Fiberstars: visual comparison of diffusion tractography data between multiple subjects." 2021 IEEE 14th Pacific Visualization Symposium (PacificVis). IEEE, 2021.
Details
- Student
-
TLTeodor Lungu
- Supervisor
-
 Maxime Chamberland
Maxime Chamberland
- Link
- Thesis