Project: Brain as a Dynamic Network Visualization: Can We See the Parts Affected by Depression?
Description
Background
Major depressive disorder (MDD) is a prevalent neuropsychiatric disorder with at least one episode occurring in the lifetime of 15%-18% of people worldwide. MDD is a very heterogeneous disease with highly varying symptom profiles and inefficient treatment as only one-third remit after first-line treatment. Identification of imaging biomarkers could improve the current standard of subjective diagnosis and prognosis of MDD. Functional MRI (fMRI) is an imaging modality that is often used to explore biomarkers in the form of activation patterns and interaction in and between brain networks, respectively. To find a biomarker, machine learning models are usually applied to classify features extracted from the fMRI scans[1]. The right features may allow a classifier to differentiate between MDD and a healthy person, but the interpretation of the features is just as important.
Goal
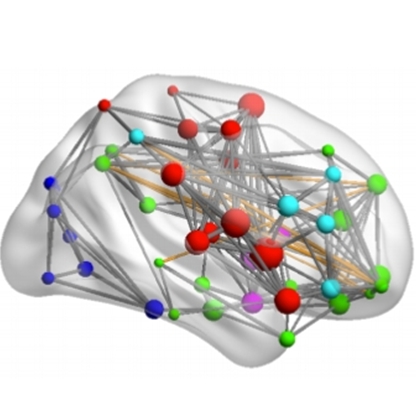
A dataset comprising 51 individuals with MDD and 21 healthy controls is available with extracted time series per functional brain network for each individual. The brain can be described as a graph in which nodes are defined by the brain regions and edges by a form of effective connectivity. It is of interest to build a dynamic graph that shows how the weights of edges change over the duration of the scan. Matlab implementations of Granger causality and wavelet coherence are available for the extraction of effective connectivity between two time series.
Classifiers are trained to achieve this task. Understanding the feature space and functioning of the model beyond derived scalar quantities can facilitates the discovery of biomarker. The goal of this project is to provide visual analytic solutions that allow the understanding of the behavior of the trained models including relevant domain information such as anatomical regions. Visualization of dynamic networks [2] in the context of cohort and explainable AI analysis is a subject that has not been extensively explored and poses many challenges that can be addressed in this master project.
This project is in collaboration with the Biomedical Diagnostics Lab at Electrical Engineering department.
Requirements
- Good programming skills
- Signal or image processing understanding
- Understanding of ML models and their inner working, ideally also explainable methods
- Visualization background
- Good communication skills
References
[1] Cîrstian, R., Pilmeyer, J., Bernas, A., Jansen, J. F. A., Breeuwer, M., Aldenkamp, A. P., & Zinger, S. (2023). Objective biomarkers of depression: A study of Granger causality and wavelet coherence in resting-state fMRI. Journal of neuroimaging : official journal of the American Society of Neuroimaging, 33(3), 404–414. https://doi.org/10.1111/jon.13085
[2] Interactive Visualization of Dynamic Multivariate Networks, Stef van den Elzen , PhD Thesis, Eindhoven University of Technology, 2015 https://research.tue.nl/nl/publications/interactive-visualization-of-dynamic-multivariate-networks
Image: Brain network visualization by Mingrui Xia, Jinhui Wang, and Yong He. Brainnet viewer: a network visualization tool for human brain connectomics. PloS one, 8(7):e68910, 2013.
Details
- Student
-
TOTim Htet Yan Oo
- Supervisor
-
 Stef van den Elzen
Stef van den Elzen
- Secondary supervisor
-
 Anna Vilanova
Anna Vilanova
- External location
- Sveta Zinger and Sjir Schielen supervisors (EE)