Project: Feature selection and ranking for Radiogenomics model of prostate cancer
Description
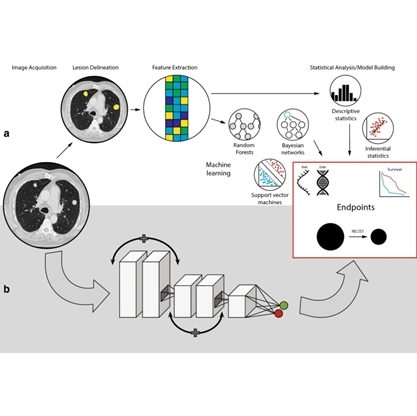
Background
Prostate cancer is a major health burden. Both in Europe and the United States, it is the type of cancer with the highest incidence in men and the second cause of cancer-related death. One in seven men is diagnosed with prostate cancer in his lifetime, but only a fraction of these tumors are aggressive and require prompt intervention. Multi-parametric magnetic resonance imaging (mpMRI) can be used to visualize tissue anatomy and physiology, and it is the recommended imaging modality for prostate cancer diagnosis. However, large multicenter studies demonstrate that mpMRI has limitations, with over 40% false positives while missing 10-15% of aggressive tumors. Recognition of cancer as a genetic disease has driven increasing research efforts towards cancer genomics, i.e., the study of the differences in DNA and gene expression between cancer and normal cells. Genomic analysis has shown a large variability in the genetic makeup of different tumors within the same prostate and even relevant differences within the same tumor region, which likely explains the large variations observed in clinical outcomes. An integrated approach, combining the strengths of both image-based and genetic analysis is necessary to enable accurate diagnosis and prognosis of prostate cancer. To this end, we have extracted a large number of imaging and genomic features from mpMRI and the RNAseq acquired on tumor biopsied lesions. In parallel, we have built a radiomics models with the imaging features and a genomic model with the genomic features to classify aggressive prostate cancer by machine learning. We then investigated the correlation between imaging and genomic features, with interesting initial finding. However, the large dimensionality of the imaging and genomic feature sets makes it challenging to understand which are the most relevant for identifying aggressive prostate cancer that are worth further investigation.
Project Objectives
In this project, we aim at developing a visual analytics methods tailored to better understand the feature importance in the machine learning decision process for prostate cancer. In other words, we aim to investigate visual analytics solutions that support identifying features or combinations of features that are most important for aggressive prostate cancer. Additionally, another visualization methods for understanding the link between imaging and genomic features need to be developed. Visual analytics systems that address these issues have been developed[1,2], however, the specific situation of imaging in combination with genomic data adds a level of complexity that as not been addressed in the past.
Requirements
- Good programming skills
- Understanding
of ML models and their inner workings, ideally also explainable methods
- Medical image analysis
- Visualization
background
References
[1] Meng, Linhao, Stef Van Den Elzen, and Anna Vilanova. "ModelWise: Interactive Model Comparison for Model Diagnosis, Improvement and Selection." Computer Graphics Forum. Vol. 41. No. 3. 2022.
[2] Raidou, Renata Georgia, et al. "Visual analytics for the exploration of tumor tissue characterization." Computer Graphics Forum. Vol. 34. No. 3. 2015.
* image from Bodalal, Z., Trebeschi, S., Nguyen-Kim, T.D.L. et al. Radiogenomics: bridging imaging and genomics. Abdom Radiol 44, 1960–1984 (2019). https://doi.org/10.1007/s00261-019-02028-w
Details
- Student
-
HKHilke Kroes
- Supervisor
-
 Anna Vilanova
Anna Vilanova
- Secondary supervisor
-
 Linhao Meng
Linhao Meng
- External location
- Simona Turco (BM/d EE)