Project: Enhancing Sequence Tube Maps for Pangenome Graph Visualization
Description
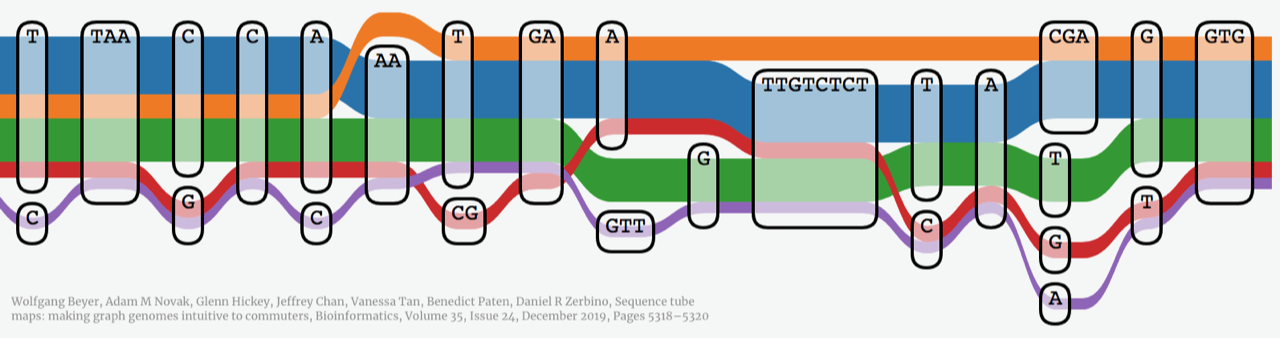
In recent years, graph pangenomes have become popular approaches to compare the DNA sequences of many organisms and discover differences that link to observable and measurable characteristics (or phenotypes) of interest, such as higher yield for crops or disease resistance in human genomics. These approaches have gained popularity over traditional linear reference comparisons because the graph makes finding similarities and differences between many sequences easier.
To interpret (small variations) the graph, users create visualizations of pangenome graph with methods such as Sequence Tube Maps [1]. This method compactly shows the paths of variation in the graph, like a subway or tube map. Although the technique provides a visually pleasing view for a relatively small region, it has readability, flexibility, and navigation limitations when considering larger regions and regions with many variations.
Goals:
In this project, you will explore methods to address some of the limitations mentioned above, specifically by:
(1) first, identifying what are the pros and cons of this technique and for which user analysis tasks (i.e., how informative is it to "look at the graph"?)
(2) considering visualization modifications to better support these tasks
(3) exploring (custom) interactions to emphasize important aspects
Requirements:
- Experience with javascript programming and d3.js
- Self-motivated to learn (or ideally already familiar with) with graph layout algorithms
- Basic knowledge of linux (and bash)
Reach out to Astrid van den Brandt (a.v.d.brandt@tue.nl) if you are interested in this project.
Reference:
[1] Wolfgang Beyer, Adam M Novak, Glenn Hickey, Jeffrey Chan, Vanessa Tan, Benedict Paten, Daniel R Zerbino, Sequence tube maps: making graph genomes intuitive to commuters, Bioinformatics, Volume 35, Issue 24, December 2019, Pages 5318–5320,
Code: https://github.com/vgteam/sequenceTubeMap
Details
- Student
-
JGJasper Geelen
- Supervisor
-
 Huub van de Wetering
Huub van de Wetering
- Secondary supervisor
-
 Astrid van den Brandt
Astrid van den Brandt